Technology
A total infrastructure of over 90 state-of-the-art mass spectrometers are available in eleven transnational access sites.
With over 30 Orbitrap’s, more than 10 Q – TOF’s, and 5 Triple Quad’s, the platform also houses a series of MALDI-TOF/TOF’s and FT-ICR high resolution instruments. Different access sites will host projects with different requirements (e.g. PTM analysis, membrane protein analysis, Bio-imaging, native structure analysis, native MS).
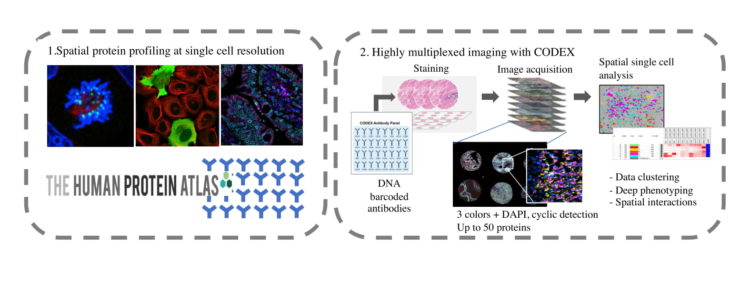
As a complement to the mass spectrometry focused sites, the KTH Access site offers antibody based imaging to study spatial proteomics in situ. The KTH access site provides single cell analysis via high-throughput imaging using a wide collection of antibodies from the Human Protein Atlas to study protein location at the single cell level or highly multiplexed imaging of 30 proteins using the CODEX platform.

 UTRECHT UNIVERSITY
UTRECHT UNIVERSITY
 UNIVERSITY OF COPENHAGEN
UNIVERSITY OF COPENHAGEN
TECHNOLOGY
Shotgun proteomics
- 2x Q-Exactive HF-X
- 1x Q-Exactive HF
- 1x Fusion Lumos
- 1x Fusion
- 1x Orbitrap-XL
- 3x triple-quad
Native / top-down proteomics
- 2x UHMR
- 8x QTOF type instruments
Sample preparation
- Agilent BRAVO
- SEC / SCX / high-pH / Mixed-phase
Software
- XlinkX in Proteome Discoverer 2.2/2.3
TECHNOLOGY
Shotgun proteomics
- 10x Q-Exactive HF-X
- 1x Orbitrap Fusion Lumos
Online LC-MS/MS
- 12x Easy nLC1200
- 2x Easy nLC1000
- 3x EvoSep OneLC
The instruments include 10 of the latest generation of Q Exactive HF-X mass spectrometers exhibiting improved speed and sensitivity, and one Orbitrap Fusion Lumos mass spectrometer providing complementary fragmentation techniques and scanning techniques, such as ETD and MS3 acquisition.
 ETH/ FUNCTIONAL GENOMICS CENTER ZURICH
ETH/ FUNCTIONAL GENOMICS CENTER ZURICH
 TECHNICAL UNIVERSITY OF MUNICH
TECHNICAL UNIVERSITY OF MUNICH
TECHNOLOGY
Mass Spec Instruments
- 1x Q-Exactive HF
- 2x Q-Exactive
- 2x Fusion Lumos
- 1x Fusion
- 1x TimsTOF pro
- 1x Triple-quad
- 1x MALDI-TOF
This facility houses nine mass spectrometers, which are equipped for nanoLC-MS/MS proteomics-type experiments.
The instruments include state-of-the-art technologies like high-resolution Orbitrap mass spectrometry, CID, HCD, electron transfer dissociation (ETD), and combinations thereof (EThcD), and cross-linking MS.
Collectively, the analytical technologies provide a full-spectrum infrastructure for high throughput and in-depth proteome analysis. Based on the broad fundament of methodological expertise and know-how, also targeted MS-based approaches, including selected reaction monitoring (SRM) and parallel reaction monitoring (PRM), as well as data independent acquisition (DIA)
TECHNOLOGY
Mass Spectrometers
- 4x Orbitrap Lumos,
- 3x Orbitrap HF-X
Expertise
- DDA (SILAC, TMT, label-free)
- DIA (PRM, SWATH)
- Bottom up proteomics
- Full proteomes
- Phosphoproteomes, PTMs
- Affinity proteomes
- Tissues, body fluids
The infrastructure boasts the latest hardware for proteomics including three Orbitrap Fusion Lumos instruments (equipped with electron-transfer dissociation, ETD or ultra-violet photo-dissociation, UVPD) and three Orbitrap Q-Exactive HF-X instruments. The Orbitrap instruments are furnished with ultra-high performance nano-liquid chromatography systems. This outstanding equipment base enables proteomic projects at any scale and ranging from high-throughput discovery (data-dependent acquisition, DDA), targeted (parallel reaction monitoring, PRM) and data-independent (DIA) experiments.

VLAAMS INSTITUUT VOOR BIOTECHNOLOGIE (VIB), GHENT, BELGIUM
 PASTEUR INSTITUTE, FRANCE
PASTEUR INSTITUTE, FRANCE
TECHNOLOGY
Mass Spec Instruments
- 6 x mass specs including several Orbitraps
- 1x Triple Quad
- 1x MALDI MS/MS
This facility houses 8 mass spectrometers, all coupled to sensitive nano-LC chromatography systems.
In combination with PSD, CID, HCD and ETD peptide fragmentation possibilities, these instruments enable high-throughput analysis of complex proteome samples as well as intact protein measurements. In addition, workflows for targeted MS-based approaches, including selected/parallel reaction monitoring (SRM/PRM) and data independent acquisition (DIA) are in place.
TECHNOLOGY
Mass Spec Instruments
- 1x Orbitrap LTQ-Velos (ETD)
- 1x Synapt G2SI & HDX Manager
- 1x Orbitrap Fusion Lumos (ETD, UVPD)
- 1x Q-Exactive HF
- 2x Q-Exactive plus
The instruments include state-of-the-art technologies like CID, HCD, Surface Induced Dissociation (SID), Electron Transfer Dissociation (ETD), and combinations thereof (EThcD), Ion Mobility Separation (IMS), cross-linking MS and H/D exchange MS (with an automated HDX platform). Ultra-violet photo-dissociation (UVPD) will be available soon.
 CENTRE FOR GENOMIC REGULATION (CRG)/
CENTRE FOR GENOMIC REGULATION (CRG)/
POMPEU FABRA UNIVERSITY (UPF), SPAIN
 SPANISH NATIONAL CANCER RESEARCH CENTRE
SPANISH NATIONAL CANCER RESEARCH CENTRE
TECHNOLOGY
Mass Spec Instruments
- 1 x Orbitrap Fusion Lumos
- 1x LTQ Orbitrap XL
- 1x LTQ ORBITRAP Velos
- 1x QTrap 5500
- 1x QTOF 5600
All mass spectrometers are equipped with nanoLC chromatographic systems for quantitative proteomics experiments, and they cover a wide range of mass analysers (orbitrap, TOF, quadrupoles, ion traps), fragmentation types (CID, HCD, ETD, and their combinations), and acquisition workflows (e.g. DDA, SRM, PRM, and DIA).
TECHNOLOGY
Mass Spec Instruments
- 2 x Qex HF-X
- 1x Qex Plus
- 1x Bruker Impact QqTOF
- 1x LTQ Orbitrap Velos
- 1x QTrap 5500
NanoLC chromatographic systems are coupled to these instruments for quantitative proteomics experiments, and they cover a wide range of mass analysers (orbitrap, TOF, quadrupoles, ion traps), fragmentation types (CID, HCD, ETD, and their combinations), and acquisition workflows (e.g. DDA, SRM, PRM, and DIA).
INSTITUTE OF MICROBIOLOGY, CZECH ACADEMY OF SCIENCES, VESTEC/MASARYK UNIVERSITY, BRNO, CZECH REPUBLIC
 CEITEC MASARYK UNIVERSITY, BRNO, CZECH REPUBLIC
CEITEC MASARYK UNIVERSITY, BRNO, CZECH REPUBLIC
TECHNOLOGY
Mass Spec Instruments
- 2x FT-ICR
These facilities jointly house two FT-ICR mass spectrometers (Bruker solariX Ultra-high Resolution Hybrid Qh-Fourier Transform Mass Spectrometers). Both 12 Tesla and 15 Tesla instruments are equipped with a MALDI/ESI dual source, ETD fragmentation module and ParaCell.
The 15 T instrument is equipped with an infrared laser for infrared multiphoton dissociation. Nano RSLC 3000 (ThermoFisher Scientific) and Agilent 1200 (Agilent Technologies) HPLC systems and PAL autosampler/robot (CTC analytics), MALDI-TOF/TOF MS (Ultraflex III, Bruker Daltonics) and IMMS Q-TOF (Synapt G2 HD, Waters)
TECHNOLOGY
Mass Spec Instruments
- 1x MALDI-TOF/TOF MS
- 1x Orbitrap Fusion Lumos
- 1x Orbitrap Elite.
LC MS/MS experiments are provided by the coupling of nano LC chromatographic systems to an Orbitrap Fusion Lumos and an Orbitrap Elite.
EUROPEAN INSTITUTE OF ONCOLOGY, ITALY
TECHNOLOGY
Mass Spec Instruments
- 1 x Q Exactive HF Orbitrap
- 1x Q Exactive Plus
Sample prep
1x OFFGEL fractionator, 3100 Agilent
This facility operates one off-line HPLC system and two dedicated ultra-high-pressure liquid chromatography (UHPLC) systems coupled with a high field hybrid quadruple-orbitrap mass spectrometer (QExactive-HF) and has additional access to an identical instrument setup is also available at the centralized Consortium (COGENTECH) of the Campus, when additional measurement is needed.
This MS platform is integrated to an ample set of consolidated biochemical and analytical protocols for histone, protein, peptide, modified peptide and sub-proteomes purification prior to MS-analysis. Standard Data-dependent acquisition (DDA) methods are normally used, but hybrid DDA-single reaction monitoring (SIM) can also be employed to specifically target modified peptides in complex matrices.
Genomic Unit
HiSeq 2000, Miseq, NovaSeq 6000, Nanostring nCounter, Biomek FX, LabChip GX; Bioanalyzer 2100, TapeStation, 10X Genomics Chromium, Covaris S220 series, to perform WGS, WES, RNAseq, ChIP-Seq, RepliSeq, Methylation, single-cells RNA analyses.
Crystallography Unit
The Crystallography Unit offers Static Light Scattering, Isothermal Titration Calorimetry, Thermofluor, Mosquito nanodispenser, Crystal Imager for protein expression in insect cells (baculovirus), Biochemical characterization of macromolecular complexes and their affinities, Crystallogenesis, X-ray diffraction data collection and structure determination.
Imaging Unit
The Imaging Unit is equipped with Fluorescence and Confocal Microscopes (including FRAP, FRET, Photoactivation, Tissue Mosaic Reconstruction), Robotized microscopy for high content and resolution image cytometry, to offer services like Basic Image acquisition and Widefield Timelapse, Tissue and Organoids Fluorescence Imaging, Live Cell Imaging (Confocal Timelapse, FRAP, Photoactivation, FRET), Deconvolution (Widefield), in addition to Training and assistance for Users
 ROYAL INSTITUTE OF TECHNOLOGY, STOCKHOLM
ROYAL INSTITUTE OF TECHNOLOGY, STOCKHOLM 

The Cell Profiling facility is a Swedish national facility located at SciLifeLab but is also open for international users via the European EPIC XS network. The facility enables researchers to access the resources and expertise from the Human Protein Atlas and the primary aim is to do full service immunofluorescence projects at moderate to high-throughput level. Based on the resource of more than 25,000 antibodies and a panel of >20 different human cell lines, this facility has a unique position to investigate subcellular spatial proteomics for human and rodent biology (service area 1). Since 2020, the facility is also offering highly multiplexed imaging (up to 30 markers) of tissue sections using the CODEX platform (service area 2).
For contact and to read more about the services offered at KTH Access site and the Cell Profiling National Facility go to: https://www.scilifelab.se/facilities/cell-profiling